LKJ Priors
27 Dec 2014Visualizing the LKJ Correlation Distribution
I use multilevel models a lot. I’ve transitioned from using software like lme4 in R or mixed in Stata, which use maximum likelihood methods, to using Bayesian software like Stan or JAGS to estimate multilevel models as Bayesian hierarchical models. When mulitlevel models have 2 or more random effects, the prior for the random effects is usually a covariance matrix. Typically we want to estimate the parameters in the covariance matrix and thus we place a (hyper)prior on the covariance matrix.
Consider a simple growth-curve model for longitudinal data.
Where is the overall intercept, is the effect of time, is a person-specific intercept, is a person-specific effect of time. The ’s are the random effects, which we typically assume follow a multivariate normal distribution:
Unfortunately, choosing a prior for the covariance matrix can be difficult and, frankly, occassionally maddening. Usually I have to use an inverse-Wishart distribution, which seems easy enough. However, they can be tricky to specify once the covariance matrix gets pretty big. Further, in software like JAGS, the multivariate normal distribution is parameterized with a precision matrix, so you have to use the Wishart distribution. Finally, it is downright hard to choose a sensible prior for a covariance matrix.
Wouldn’t it be great if you could choose a prior using a correlation matrix? I, and I suspsect most folks, are a little more comfortable thinking about correlations than covariances. Fortunately, the developers of the open-source Bayesian modeling program Stan have made choosing a prior for a correlation matrix reasonably straightforward by using the LKJ Correlation Density.
It turns out it isn’t too difficult to choose a prior for this type of model because there is just a single covariance to deal with. However, suppose we extend the model above to include a random intercept and two random slopes and we estimate the covariances between all three. A possible way to set up this model in Stan is to choose whatever prior you’d like for the variances and then use the LKJ Correlation Distribution to to provide a prior for the correlations. You would then obtain the covariances by using the appropriate variances and correlation. I’ll do another post that provides the specifics.
There is only a single parameter for the LKJ Distribution - . When I first started using the LKJ Correlation Distribution, I didn’t know what to set to and the Stan documentation didn’t initially make a lot of sense to me. Fortunately, Stan provides a random number generator for the LKJ Correlation Distribution, so it is easy to visualize the distribution using random draws for various values of . The primary purpose of this post is to show how different values of affect the probability of different correlations.
What the Stan Manual Says
The Stan Manual provides the following guidance regarding (page 385, Stan Manual 2.5):
The shape parameter can be interpreted like the shape parameter of a symmetric beta distribution.
- if = 1, then the density is uniform over all correlation matrices of a given order;
- if > 1, the identity matrix is the modal correlation matrix, with sharper peaks in the density around the identity matrix for larger ; and
- if 0 < < 1, the density has a trough at the identity matrix.
Using Simulation to Visualize What the Manual Means
To see what this looks like we are going to simulate correlation matrices for = .5, 1, 2, 5, 10, and 50. We’ll then plot the density of one correlation from each of the simulated matrices to see how they look. Stan requires that you input some data, so I simulated some data out of poisson distribution to fit in Stan. We don’t care about those and they could be anything.
library(rstan)
library(reshape2)
library(ggplot2)
set.seed(123)
sim_data <- list(x = rpois(30, 5), N = 30, R = 3) First, I load the required libraries for this post. Then I set the seed and create the data needed to pass into Stan. is 30 random draws from a poisson with a rate parameter = 5, is the sample size for the poisson data, and is the number of correlations we are dealing with (recall we have the correlations among the random intercept and 2 random slopes).
Stan Model
Next I write the Stan model. The key part of this code is generated quantities block. This block creates 6 correlation matrices named Omega. The number after Omega describes the size of in the LKJ Distribution. This block also takes random draws from the LKJ Distribution with values equal to .9, 1, 2, 5, 10, or 50. We can then use these random draws to visualize the shape of the priors for a given correlation.
sim_stan <- "
data {
int<lower=0> N; // number of observations
int<lower=0> x[N]; // outcome variable
int R;
}
parameters {
real lambda;
}
model {
x ~ poisson_log(lambda);
}
generated quantities {
corr_matrix[R] Omega0;
corr_matrix[R] Omega1;
corr_matrix[R] Omega2;
corr_matrix[R] Omega5;
corr_matrix[R] Omega10;
corr_matrix[R] Omega50;
Omega0 <- lkj_corr_rng(R,.9);
Omega1 <- lkj_corr_rng(R,1);
Omega2 <- lkj_corr_rng(R,2);
Omega5 <- lkj_corr_rng(R,5);
Omega10 <- lkj_corr_rng(R,10);
Omega50 <- lkj_corr_rng(R,50);
}
"Using rstan to fit the model
Next, I fit the model in R through the rstan package.
sim_parms <- c('Omega0', 'Omega1', 'Omega2',
'Omega5','Omega10', 'Omega50')
fit_sim <- stan(model_code = sim_stan, pars = sim_parms,
data = sim_data, chains = 1, iter = 10000)Processing the MCMC draws and getting them prepped for ggplot
Finally, I process the data and transform it so that I can plot the density in ggplot2.
res_sim <- as.data.frame(fit_sim)
names(res_sim) <- sub("\\[", "_", names(res_sim))
names(res_sim) <- sub("\\,", "_", names(res_sim))
names(res_sim) <- sub("\\]", "", names(res_sim))
smalldata <- res_sim[, c("Omega50_3_1", "Omega10_3_1", "Omega5_3_1",
"Omega2_3_1", "Omega1_3_1", "Omega0_3_1")]
smalldata$draw <- seq(1:length(smalldata$Omega50_3_1))
plotdata <- melt(smalldata, id = c("draw"))
plotdata$eta <- factor(plotdata$variable,
levels = levels(plotdata$variable),
label = c("eta = 50", "eta = 10",
"eta = 5", "eta = 2",
"eta = 1", "eta = .9"))
my.labs <- list(bquote(eta == 50), bquote(eta == 10),
bquote(eta == 5), bquote(eta == 2),
bquote(eta == 1), bquote(eta == .9))Creating the Plot
Here’s the code for the plot.
p <- ggplot(plotdata, aes(x = value, colour = eta))
p + geom_density() +
scale_colour_manual(values=1:6, breaks = levels(plotdata$eta),
labels = my.labs, name = "Shape") +
xlab("Correlation Value") +
ylab("Density") +
ggtitle("Visualization of a correlation from the
lkj_corr density in Stan \n for various
values of the shape parameter") +
theme_bw()Visualizing the Density
The moment you’ve been waiting for – a plot of the density.
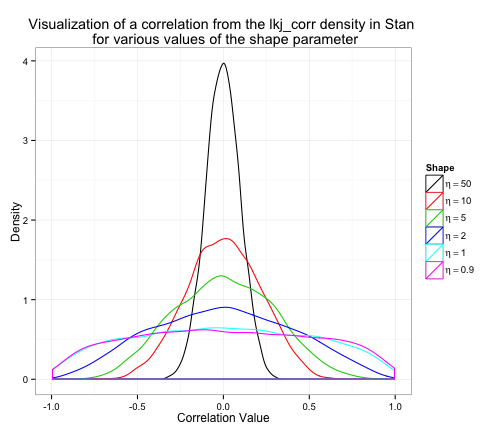
These plots are consistent with what the Stan manual says:
The shape parameter can be interpreted like the shape parameter of a symmetric beta distribution.
- if = 1, then the density is uniform over all correlation matrices of a given order;
- if > 1, the identity matrix is the modal correlation matrix, with sharper peaks in the density around the identity matrix for larger ; and
- if 0 < < 1, the density has a trough at the identity matrix.
I find this helps me make better decisions about the value of .